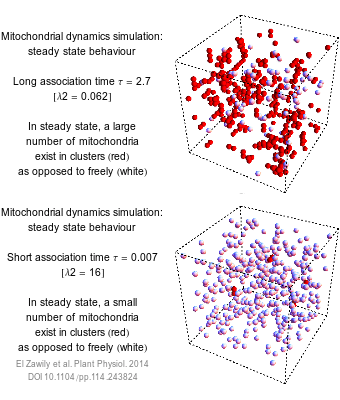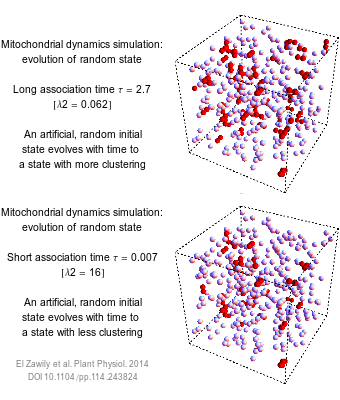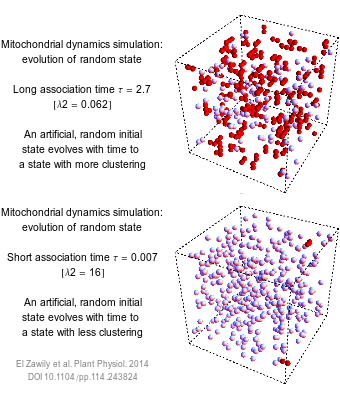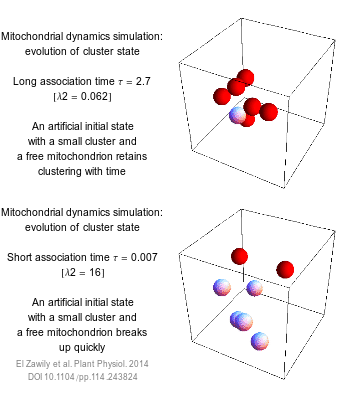
The numbers that we use to describe the world are rarely exact. How long
will it take you to drive to work? Perhaps "between 20 and 30 minutes".
It would be unwise (and unnecessary) to say "exactly 23.4 minutes".
This uncertainty means that "back-of-the-envelope" calculations are very
valuable in estimating and reasoning about numerical problems,
particularly in the sciences. The idea here is to perform a calculation
using rough guesses of the quantities involved, to
get an "order of magnitude" estimate of the answer you're after. Made
famous in physics as "Fermi problems", attributed to Enrico Fermi (who
used rough reasoning to deduce quantities from the power of an atomic
bomb to the number of piano tuners in Chicago),
this approach is integral in many current applications of maths and
science. Cool books like "Street-fighting Mathematics", "Guesstimation", "Back of the envelope physics", the excellent
"What If?" section of xkcd, and the lateral interview questions facing some job candidates: "how much of the world's water
is contained in a cow?" are all examples.
We've built a free online calculator (Caladis -- calculate a
distribution) that complements this approach by allowing one to take the
uncertainty in one's estimates into account throughout a calculation.
For example, what volume of CO2 is produced by our yearly
driving? We could say that we cover 8000 miles per year "give or take"
1000 miles, and find that our car's CO2 emissions are between 100 and
150 grams per kilometre. Our calculator allows us to do the necessary
conversions and sums while taking this possible
variability into account -- doing maths with "probability
distributions" describing our uncertainty. We no longer obtain a single
(possibly inaccurate) answer, but a distribution telling us how likely
any particular answer is -- in this case a rather concerning
bell-shaped distribution between 1 and 2 tonnes which can be viewed here
In the sciences, particularly in biology, measurements often have
substantial uncertainties -- due to experimental error, natural
variability in the system of interest, or both -- and so using
distributions rather than single numbers in calculations allows
us to understand and process more about the question of interest. "Back-of-the-envelope" calculations are certainly useful in biology but, owing to the uncertainties involved, one can trust one's estimates better if one has a smart envelope that takes that uncertainty into account. We've
written an accompanying paper "Explicit tracking of uncertainty increases the power of quantitative rule-of-thumb reasoning in cell biology" (free to all in Biophysical Journal) showing how to use our calculator -- in
conjunction with the excellent Bionumbers online database, a collection
of (often uncertain) experimental measurements
in biology -- to make real biological calculations more powerful. Do have a go at using our calculator at
www.caladis.org : it's user-friendly and there are lots of examples showing how it works! Iain and Nick